God’s Wisdom in the Genome
The genome of organisms contains their genetic material, which largely determines how they will develop. Scientists are learning more and more about the genome of various organisms, which is revealing more and more about the amazing wisdom of God in their design and creation. It seems that our understanding of the functioning of life is always incomplete, and the genome is always more complex than we had thought. Will we ever fully understand the functioning of life, or will it always be a mystery to us?
First we present excerpt from an article about genetics in a recent issue of a magazine produced by the University of North Carolina at Chapel Hill. This article shows some unexpected features of the genome. What one typically calls a gene is really a protein-coding gene and contains instructions for making one or more proteins. The DNA that is not included in such genes has been called “junk DNA” in the past, but now we are learning that this DNA has a function and contains genes that do not code for protein. This DNA that does not code for protein is now called noncoding DNA.
Humans are strikingly similar to a bunch of different critters—genetically, at least. Sixty percent of human genes are fundamentally the same as fruit fly genes, and somewhere around ninety percent of our genes are the same as mouse genes. 1
You’ve also got something called noncoding DNA [in addition to genes that code for protein]. About six feet of DNA are wrapped up in each of your cells. We used to think that only about one inch of it actually did anything; the other five feet eleven inches seemed to be junk. Now scientists are starting to think that our one inch of working DNA, along with all the proteins it produces—and RNA, and some other cellular cousins that we don’t have time to get into today—are just our nuts and bolts, and that what we once called junk DNA actually holds our blueprints and the systems that control how we’re built. 1
Thus the way the organism is organized and constructed is specified by the noncoding DNA; the protein coding genes tell the building blocks out of which the organism is made. This shows that organisms with very different structure can have similar protein coding genes because they are made of similar proteins, but the proteins are structured in a very different way. So just the differences between the genes that code for protein do not give a good idea of how similar two organisms are. Thus the one to two percent difference between humans and apes in their protein coding genes is not a good measure of how similar we are.
So it’s starting to look like genes are a lot more complicated than we thought. A given gene may do a whole lot more than make one protein or control one process. Scientists once speculated that we’d need at least a hundred thousand genes to build a human. But we’ve only got about twenty-two thousand. Some worms have more than that. A lungfish has forty times more DNA than you or I have. In a given gene, different combinations of T 2 coding DNA can become active at different times to produce different proteins, and that helps explain how humans can be so complex with relatively few genes. But we still have no idea how our cells “know” which parts of a gene to pay attention to at any given time, and we really don’t know what determines which genes get switched on or off at specific times and in specific places. 1
Another big point: we don’t need a whole gaggle of genes to make new stuff. For example, a single gene called BMP4 helps determine which parts of an embryo will become the back and belly. But BMP4 also determines whether a finch has a broad beak or a long one. It also specifies whether a cichlid fish is going to have a short, thick jaw for crushing mollusks, or a longer one for sucking algae. One gene, switched on at different times and places, may build flippers in whales and arms in football players. 1
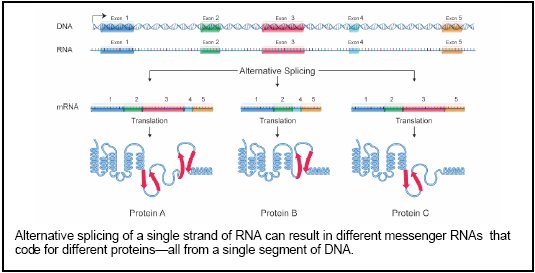
But before cells can produce proteins, they have to transcribe DNA into something called premessenger RNA. Pre-messenger RNA molecules are then spliced—little sections are cut out and then recombined in different ways—to create mature messenger RNA. This mature stuff then gets translated into a protein. One RNA can be spliced several different ways to yield different proteins, and different variants of the same protein are often found in different tissues, where their subtly different sequences allow them to do specific jobs. In fact, it’s impossible for us to count how many different proteins there are in humans. And if all that isn’t complicated enough, sometimes the little buggers pair up and work in teams. 1
Remember the non-coding DNA we talked about earlier, the stuff that scientists once thought was junk? Did I mention that it makes up about 99 percent of our DNA? We’re learning how to read it, but we’ve just started. 1
The next article presents more information about the noncoding DNA and shows that even the DNA that differs between closely related species has a function. This article discusses how scientists have determined that a human chromosome put in a mouse cell still functions largely as it does in humans. 2 This is despite the fact that human and mouse protein coding genes are very similar. Thus it is gene regulation and the noncoding DNA that greatly differs between humans and mice that determines gene expression, not the protein coding genes themselves. If the protein coding genes determined the function of the DNA, then the human chromosome would function nearly the same as a mouse chromosome because the protein coding genes are so similar.
The noncoding sequences differ greatly between human and mouse. These are the parts that determine how genes are expressed (turned on and off) in a cell. Thus even parts of the DNA that differ greatly between species can be functional. This raises the question how such parts of the DNA could have changed so rapidly between human and mouse according to the theory of evolution and still be functional when the genes themselves changed little. It also shows that much more of the DNA is functional than we had thought.
Referring to a paper by Wilson et al., 3 the authors state, “The paper’s findings also call into question one of the basic tenets of comparative genomics: that evolutionary conservation can serve as the primary tool for finding functional sequences.” 2
This conclusion seems to raise serious problems for the theory of evolution. Where did all this species-specific DNA, which is even functional, come from? How could it have evolved, and evolved so quickly, under evolutionary assumptions if it is so different between humans and mice?
The third article shows that a very large percentage of the genome is translated into RNA, indicating that what was once considered to be “junk” DNA has some kind of a function.
According to a painstaking new analysis of 1% of the human genome, genes can be sprawling, with far-flung protein-coding and regulatory regions that overlap with other genes. 4
As part of the Encyclopedia of DNA Elements (ENCODE) project, 35 research teams have analyzed 44 regions of the human genome covering 30 million bases and figured out how each base contributes to overall genome function. The results…provide a litany of new insights and drive home how complex our genetic code really is. For example, proteincoding DNA makes up barely 2% of the overall genome, yet 80% of the bases studied showed signs of being expressed. 4
Given the traditional gene-centric perspective, that finding “is going to be very disturbing to some people,” says John Greally, a molecular biologist at Albert Einstein College of Medicine in New York City. On the other hand, says Francis Collins, director of the National Human Genome Research Institute (NHGRI) in Bethesda, Maryland, “we’re beginning to understand the ground rules by which the genome functions.” 4
“A lot more of the DNA [is] turning up in RNA than most people would have predicted,” says Collins. 4
But in the 8 June issue of Science (p. 1484), Gingeras and his colleagues reported that many of the mysterious RNA transcripts found as part of ENCODE harbor short sequences, conserved across mice and humans, that are likely important in gene regulation. That these transcripts are “so diverse and prevalent across the genome just opens up the complexity of this whole system,” says Gingeras. 4
The mRNA [messenger RNA] produced from protein-coding genes also held surprises. …they found additional exons—the regions that code for amino acids—for more than 80% [of the protein coding genes]. Many of these newfound exons were located thousands of bases away from the gene’s previously known exons, sometimes hidden in another gene. Moreover, some mRNAs were derived from exons belonging to two genes, a finding, says Reymond, that “underscores that we have still not truly answered the question, ‘What is a gene?’ “ In addition, further extending and blurring gene boundaries, ENCODE uncovered a slew of novel “start sites” for genes--the DNA sequences where transcription begins--many located hundreds of thousands of bases away from the known start sites. 4
The distributions of exons, promoters, gene start sites, and other DNA features and the existence of widespread transcription suggest that a multidimensional network regulates gene expression. 4
The next article gives additional evidence that the great majority of the genome is transcribed into RNA, and therefore has a function.
…a substantial amount of the genome outside the boundaries of known or predicted genes is transcribed. 5
…non-genic transcription is very widespread. Precisely how wide is not yet known. Moreover, the function of this non-genic, transcribed material is unclear. 5
…some of this [transcription] activity occurs in pseudogenes, regions of the genome long considered fossils of past genes. In a transcriptional sense, dead genes appear to come to life, with some clues even suggesting that they may help regulate other genes. 5
When a sequence from outside the conventional bounds of a gene is spliced in as well, the number of variants increases further. …This variation [alternative splicing] too appears to be considerably more prevalent than once was thought. 5
…genes can be regulated very far upstream by enhancers over 50,000 base pairs away, even beyond adjacent genes. The looping and folding of DNA can bring distant spans into close spatial proximity. 5
Attachment of methyl groups to DNA and modification of histones that support the DNA can also affect gene activity.
…the recent ENCODE results suggest that deviations from the traditional model [of genes] could be the norm. 5
The next article gives evidence that transposons, also once thought to be without a function, also have a function. It shows that transposons are often similar and in similar locations in different organisms. Even under creationary assumptions, this indicates that these transposons have a function.
…Researchers sifting through the supposed junk DNA between genes—a whopping 98% of the human genome—have in the past few years hit a mother lode of functional sequences full of clues about how genomes operate and change through time. And, as junk DNA has gained respect, so have mobile bits of DNA called transposons that are often the source of this genomic clutter. 6
Most researchers have taken a dim view of transposons, considering them molecular parasites that clog chromosomes with seemingly useless sequence, sometimes disrupting genes. Now, comparative surveys, along with experimental studies of gene regulation, are showing that transposons can influence when, where, and how genes are expressed. 6
Transposons, small packages of DNA that can splice into other sequences, seem to appear suddenly in a genome, copying, cutting, and pasting themselves throughout its chromosomes. …A few…are found in the same place in the genomes of many species. To be so highly conserved, they must play a role so important to survival that evolution keeps them intact, weeding out deleterious mutations. 6
In 2004, Bejerano, then at the University of California, Santa Cruz, and his colleagues described more than 400 stretches of at least 200 bases that were the same in human, rat, mouse, chicken, dog, and, to a lesser extent, fish. Three-quarters of these so-called ultra-conserved regions resided outside genes (Science, 28 May 2004, p. 1321). Last year, Byrappa Venkatesh of the Institute of Molecular and Cell Biology in Singapore and colleagues compared DNA of elephant sharks and humans,…Working with a very sketchy draft of the shark genome, they found 4800 conserved sequences. Like others, they found that these conserved sequences tended to cluster near genes for proteins that regulate transcription and DNA binding. 6
Gradually, researchers began to realize that some of these conserved elements [between elephant, shark, and humans] were transposons. The coelacanth, a “living fossil” species that [supposedly] dates back more than 400 million years, led Norihiro Okada of the Tokyo Institute of Technology to a whole conserved superfamily of transposons called short interspersed repetitive elements (SINEs). Okada and his colleagues first found two SINEs in the coelacanth whose sequences looked similar enough to each other to have a common origin; they then searched for the same sequences in genome databases. They found them [conserved transposons] in salmon, trout, hagfish, dogfish sharks, lancelets, catfish, zebrafish, and the sea urchin….Yet another analysis unearthed 1000 copies of a subset of these SINEs [transposons], called AmnSINEs, in both humans and chickens. 6
When Broad Institute researchers Xiaohui Xie and Michael Kamal trekked through the human genome looking for stretches of DNA that occurred multiple times, they found one that looked quite a bit like the core of a zebrafish transposable element, also part of the SINE family. Eventually, they turned up 123 more copies, some more complete than others. They found this SINE’s 180-base core in the same places in other genomes and a few copies in the coelacanth. 6
When Bejerano took a close look at his ultraconserved sequences, he too discovered the remains of a family of transposable elements. Through genome comparisons, he [Bejerano] and his colleagues found LF-SINE [“lobe-finned” fish transposon] variations in human, chicken, dog, and all the other tetrapod sequences in the public databases. 6
The conservation of the sequence in similar places in the genomes of all these species suggested they play a key role in genome function. Bejerano tested one that was located 500,000 bases from a gene coding for a transcription factor active in motor neuron development. [He found evidence that it does help to regulate the gene.] 6
Now work by Bejerano and others has shown that functional transposable elements are more than a fluke. [They looked at the opossum genome and other placental mammals’ genomes.] 6
Overall, Bejerano and his colleagues have just found more than 10,000 conserved transposons in the human genome.…He and his colleagues first identified these sequences by looking for conserved stretches across a range of vertebrate genomes, including human, and subtracting out any that represented genes. 6
At this point, the evidence for a role for most of these partially preserved transposons is circumstantial. Their conservation suggests they have a function; otherwise, they should slowly disappear. 6
Scientists are also discovering many differences between humans and apes, both in their genomes and in the structure of the brain. 7 ,8 Thus the often quoted 1 to 2 percent difference between humans and apes is now not considered an accurate picture of the total difference, and the real difference, especially in higher cognitive functions, is much larger.
From the creationary viewpoint, one would only expect almost all of the DNA to be functional, and science is bearing this out. Also, since God’s wisdom is far above ours, it is only natural that the functioning of life and of the genome would still be beyond our understanding. Furthermore, because man is made in the image of God, one would expect humans and apes to differ significantly. Surely the depth and complexity of the human genome reinforces the Biblical picture of the Creator God as infinite in wisdom and power and worthy of our worship and praise.
Bible References
Psalm 139 (14) I will praise you; for I am fearfully and wonderfully made: marvelous are your works; and that my soul knows right well. (15) My substance was not hid from you, when I was made in secret, and carefully formed in the depths of the earth. (16) Your eyes saw my substance, yet being imperfect; and in your book all my members were written, which continually were fashioned, when as yet there were none of them.
Psalm 8 (4) What is man, that you are mindful of him? and the son of man, that you visit him? (5) For you have made him a little lower than the angels, and have crowned him with glory and honor.
Genesis 1 (27) So God created man in his own image, in the image of God created he him; male and female created he them.
Psalm 139 (5) You have beset me behind and before, and laid your hand upon me. (6) Such knowledge is too wonderful for me; it is high, I cannot attain to it.
- 1 a b c d e f Smith J (2008) What Don’t We Know, Endeavors 24(3). Also 2008 May 14 Accessed 2008 Nov 15
- 2 a b Coller HA, Kruglyak L (2008) It’s the Sequence, Stupid, Science 322(5900):380-381
- 3Wilson MD, Barbosa-Morais NL, Schmidt D, Conboy CM, Vanes L, Tybulewicz VL, Fisher EM, Tavaré S, Odom DT (2008) Species-Specific Transcription in Mice Carrying Human Chromosome 21, 322(5900):434-438
- 4 a b c d e f g Pennisi E (2007) DNA Study Forces Rethink of What it Means to Be a Gene, Science 16(5831):1556-1557
- 5 a b c d e f Seringhaus M, Gerstein M (2008) Genomics Confounds Gene Classification, Am Sci 96(6):466-473.
- 6 a b c d e f g h i j k Pennisi E (2008) Jumping Genes Hop Into the Evolutionary Limelight, Science 317(5840):894-895.
- 7Cohen J (2007) Relative Differences: The Myth of 1%, Science 316(5833):1836
- 8Balter M (2007) Neuroanatomy. Brain Evolution Studies Go Micro, Science 315(5816):1208-11