Recent Discoveries in Genetics
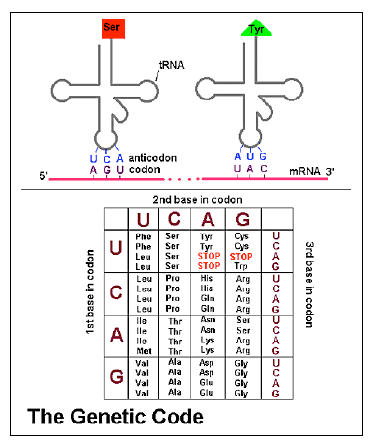
Figure 1
Several recent discoveries in genetics reveal even more of the amazing properties of the genome and give additional evidence for the design of life by an intelligent creator. DNA consists of a long sequence of four nucleotide bases. Proteins consist of sequences of about twenty amino acids, and are specified by a coding system in which three successive nucleotide bases of DNA constitute a “codon” and are translated into a specific amino acid. However, there are more codons than amino acids, so more than one codon corresponds to a given amino acid in many cases.
Now, within everyone’s DNA are “SNPs”, single nucleotide polymorphisms. This is a site at which many humans have differing nucleotides. Sometimes such a difference does not influence the amino acid coded for. For example, the codons UCU and UCC both code for serine, so a change of uracil to cytosine at this point in the RNA, corresponding to a change of thymine to cytosine in the DNA, has no effect on the sequence of amino acids in the protein. Is there then any difference between such codons in terms of their effects on the organism?
Recent research shows that there is. 1 This research shows that a combination of three SNP’s in a gene known as the MDR1 gene alters P-glycoprotein activity among individuals with these polymorphisms. Pglycoprotein is a “multiple-transmembrane protein pump” that transports various drugs out of cells. Thus this protein can affect the degree to which many drugs are absorbed and can thereby affect their utility. Therefore the combination of these three SNP’s, although it does not affect the sequence of amino acids in this protein, does affect its three-dimensional shape, and therefore the activity. How can this be?
When a protein is created from a gene, the gene is first transcribed into messenger RNA that travels to the ribosome, where the messenger RNA is translated into protein, one codon, one amino acid at a time. As this is done, a sequence of amino acids emerges from the ribosome and gradually folds up into the protein as it is is being created. The speed at which the amino acid sequence is created can affect the way in which the protein folds. It turns out that some codons are translated faster than others and this can affect the way the protein folds up. This is partly because the translation into protein depends on the availability of “transfer RNAs” (tRNAs) to carry amino acids to the ribosome. Some tRNAs are more abundant in a cell than others. Each codon specifies a different tRNA, so even if two codons specify the same amino acid, one codon may translate faster if its tRNA is more abundant.
This research was done on HeLa cells as well as cells from two species of monkeys and another line of human cells, with similar results. HeLa cells are an immortal cell line used in medical research. The cell line was derived from cervical cancer cells taken from Henrietta Lacks, who died from her cancer in 1951. These cells are called immortal because they can divide an unlimited number of times as long as they are maintained and sustained in a suitable environment. They are immortal because their telomerase is active during division so that the telomeres at the ends of the DNA do not shorten with time as in typical human cells. HeLa cells have a typical chromosome number of 82, with four copies of chromosome 12 and three copies of chromosomes 6, 8, and 17. They have become a laboratory "weed" and sometimes contaminate other cell cultures in the same laboratory, interfering with biological research. However, they are useful for research purposes because they still have many of the properties of human cells. Because this research was done on several different kinds of cells, its conclusions have more weight than if they were just done on one line of cells.
This research shows that the extra codons in the genetic code are not “wasted” and have a function in protein folding. Certain codons that take longer to translate give the protein a little more time to fold before new amino acids are produced. This can help the protein to fold properly. This is just one example of the amazing functionality that is built into many aspects of living organisms. This also helps to explain why different organisms have a different “codon bias,” a different preference for various codons that code for the same protein.
Another function built into the DNA of bacteria is described by Marx. 2 These experiments were performed on the bacterium Streptococcus thermophilus, which is widely used to make yoghurt and cheese. Bacteria are often attacked by viruses called bacteriophages, and these viruses are very abundant on earth. This research involves a defense mechanism of bacteria against these viruses. This defense mechanism involves the so-called CRISPR sequences, which are “clustered interspaced short palindromic repeats” and are widely distributed in the genomes of both bacteria and archaea, another division of single-celled organisms lacking nuclei. Accompanying these CRISPR sequences are some cas (CRISPR-associated) genes. It now appears that when they face infection by bacteriophages, bacteria can incorporate some of the sequences of the phage DNA into their CRISPR spacer sequences. Bacteria which do this have enhanced resistance to bacteriophages. Both the spacer sequences and the cas genes have been shown to be necessary for this resistance. When such sequences were artificially inserted into bacteria, their resistance to bacteriophages increased. When such sequences were removed, their resistance decreased. Also, when faced with infection by bacteriophages, some of the bacteria incorporated such sequences that had not been present before. Inactivating the cas genes reduces this resistance.
Apparently these bacteria produce short RNA sequences from these CRISPR sequences and this RNA binds to complementary sequences in messenger RNA made by invading phages, preventing the invading phages from manufacturing protein and thus preventing them from reproducing. This work may have applications in selecting bacteria that already are resistant to bacteriophages, but it also reveals the amazing ability of these tiny organisms to modify their own DNA to become resistant to infection. And this resistance passes down to their offspring because the DNA, CRISPR sequences and all, is copied during reproduction. This shows that DNA is not static but it is dynamic and can be modified by the organism. This can be seen as another facet of the functionality built into life by the Designer. So many different elements have to work together to make this system work that it is difficult to imagine how it could have evolved. It would be interesting to know whether higher organisms have a similar ability to modify their DNA in a manner that is passed down to their offspring.
Another amazing fact about the genome is that even though only a tiny amount of the genome is occupied by protein coding genes, the great majority of the human genome is transcribed into RNA. 3 In fact, millions of distinct transcribed RNA sequences are known for the human genome. 4 These sequences are produced by the so-called RNA genes. Now, a single RNA gene can produce more than one distinct sequence. However, even after clustering these sequences according to their similarity, one compilation contains over 800,000 distinct clusters! 4 An average of 10 percent of the genome is included in such sequences. 5 This could mean that there are 800,000 or even millions of RNA genes in the human genome. Because these RNA genes can be very short, they do not occupy a large amount of the genome. However, the RNA produced by these genes apparently has a function in regulating the protein-producing genes. Some of these genes produce RNA that binds to messenger RNA and prevents the messenger RNA from being translated into protein. Thus these genes appear to play a role in gene regulation. Each such RNA gene is thought to regulate multiple protein producing genes, so the RNA genes can have a significant impact on the organism. There is even a possibility that small RNA genes may activate protein coding genes instead of only repressing them. 6 Even though the human genome is thought to have only about 20,000 to 25,000 protein coding genes, the complexity of the genome and the human organism may be much larger than this indicates because of the possibility that there are millions of small RNA genes that regulate the activity of the protein coding genes. Such great complexity is additional evidence of the thought that went into the design of life and makes it even harder to imagine how life could evolve by blind mutations.
It turns out that DNA can be transferred between even distantly related species of bacteria. In fact, according to Fraser et al. “it is probably possible, through a series of intermediaries, to transfer genes between any two bacteria”. 7 This transfer is more frequent between closely related bacteria than between distantly related ones, because of mechanisms in the bacteria that prevent a transfer if the DNA is mismatched. In spite of this, “far from a continuum mediated by promiscuous gene exchange, bacteria seem to form clusters of genetically related strains (species), at least for those genera studied so far.” The question arises as to why there are still identifiable groups of bacteria if gene exchange is so frequent, instead of a continuous variation between different kinds of bacteria. This seems to be an evidence that the barriers between different kinds of bacteria are so strong that even when evolution is helped by receiving fully functional genes from closely related species of bacteria, evolution cannot cross these barriers. Of course, the barriers between higher organisms would be even stronger, and the transfer of genetic material is less frequent, suggesting that evolution never could cross these barriers between different kinds of organisms and that instead of evolving from a common ancestor, the major kinds of life were created much as they are today.
The final item that relates to genetics has to do with the variations in the genomes of different humans. Scientists have recently developed an enhanced map of the genome that shows differences between the DNA of different members of the human population. 8 However, the scientists report that “the genetic differences in our individual DNA are greater than originally thought. For example, it is not uncommon to find in some people large sections of DNA moved to a different location, or missing altogether.” This observation is interesting for two reasons. First, it shows that our genome is so well constructed that the human organism can still function when large portions of the DNA are moved to a different location or missing altogether. This is an additional evidence of the marvelous forethought that went into the design of life, especially human life. Second, this shows that, instead of evolving to greater and greater fitness, the human organism is rapidly degenerating. This contradicts the teaching of the theory of evolution, which states that all of life developed from a simple, common ancestor by gradual changes. Instead of developing to greater and greater complexity, life is rapidly degenerating and deteriorating.
The more we learn about life and about the genome, the more we understand its amazing complexity and greater and greater is the doubt cast on the hypothesis that life evolved by random mutations and selection from nonliving matter on earth without the intervention of an intelligent Creator. If it were not for the dogmatic stance taken by the scientific community against creation and in favor of evolution, no doubt many more people would see that life was designed by a loving and wise Creator.
- 1Komar A (2007) SNPs, silent but not invisible. Science 315 (5811): 466-467
- 2Marx J (2007) New bacterial defense against phage invaders identified. Science 315:1650
- 3Mattick J (2005) The functional genomics of noncoding RNA. Science 309 (5740):1527-1528
- 4 a b Nekrutenko A (2004) Reconciling the numbers: ESTs versus protein-coding genes, Mol. Biol. Evol. 21(7):1278-1282
- 5Claverie J (2005) Fewer Genes, More Noncoding RNA, Science 309 (5740):1529-1530
- 6Garber K (2006) Small RNAs reveal an activating side. Science 314 (5800):741
- 7Fraser C, Hanag WP, Spratt BG (2007) Recombination and the nature of bacterial speciation. Science 315: 476-480
- 8Rice S, Feig C, Fortin J, Langmaid T (2006 Nov 24) Enhanced genome map could help disease research, scientists say. http://www.cnn.com/2006/HEALTH/11/24/genome.disease/index.html Accessed 2007 Apr 15