The Discovery and Implications of Orphan Genes
The goal of this paper is to review some significant findings from the analysis of genetic sequence data over the last few decades. These results have surprised many biologists and are believed by many to challenge the key assumptions about how life came to exist on this planet. In particular, these recent findings indicate that the diversity of genetic information across the entire range of life forms is much greater than previously known and that this information in the form of protein-coding genes shows that all forms of life are much more distinct from each other at a very fundamental level than ever was previously understood. These results call into question specific key assumptions that make up the evolutionary explanation for life:
- The assumption that all life forms descend from one single, simple original form of life, that is, Universal Common Descent (UCD) and
- The assumption that all life forms are related through this shared common descent as organisms evolved from very simple, single simple organism to much more complex structure and functions via naturalistic mechanisms of genetic change over time.
As we will see, instead of reinforcing evolutionary assumptions about origin and development of life, these results better fit that the conclusion that all life was specially created with intrinsic complexity and with a wide variety of distinct forms and functions.
Introduction
To understand why these results from the last several decades are so significant, we must understand that advocates of evolution believe that similarity, or homology, is the main point of evidence, or essentially the proof, of their doctrine that UCD with modification is the explanation for all life on earth. Although homology originally indicated just similarity, evolutionists now take the word to mean similarity that is a result of decent from a common ancestral organism. 1
Despite this claim that similarity proves evolution, most people understand that high levels of similarity between different organisms is also completely consistent with the reality that life was intentionally designed using similar design patterns for many different forms of life. Many different organisms despite having varied form and function still need to exist in similar environments, eat similar foods, and live together on this planet. They clearly benefit from these widely observed common design patterns. This, in fact, is essentially a universal feature of all forms of life on earth—they are like other forms yet different as well.
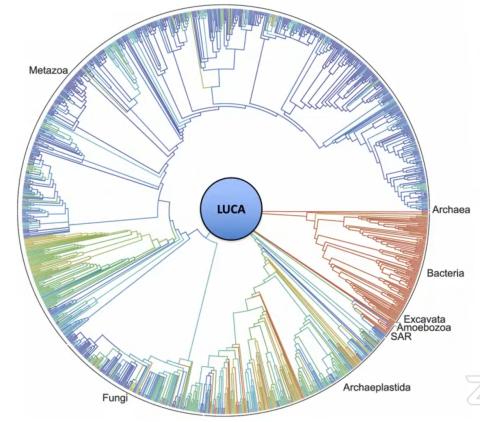
The theory of evolution assumes that this homology is due to UCD—simple to complex evolution from a single shared common “ancestor” that they now refer to as the last universal common ancestor (LUCA). Figure 1 shows one form of a phylogenetic tree that also includes this hypothetical LUCA organism at the central root of the tree.
Discovery of orphan genes and the implications for common descent
Previously, the assumed evolutionary relationships between different species or groups of organisms as shown in such phylogenetic trees were inferred from similarities in anatomy and physiology. More recently, biologists have been able to reliably recover the complete sequence of genetic material that is contained within the cells of living organisms. Although they may not fully understand the function of the entire genetic sequence and the complex regulatory systems of organisms, biologists are able to identify the genes—those shorter sequences of hundreds or thousands of base pairs along the DNA strand that translate specifically into amino acid sequences that are then folded into proteins. It is these relatively short but numerous genes that define individual specific proteins that are used in the structure and function of living organisms.
This isolation of the individual genes and comparison across different species has enabled the development of mathematical techniques intended to infer relationships between different organisms. These new techniques are based not on similar anatomy or form, but rather are derived from similarities and differences in the sequences that define proteins within the organism itself at the genetic level.
Each species or group of organisms has a unique combination of genes—some may define proteins that are common across many species, but many can be unique to a single species or group even if they serve similar roles as different proteins in other species. In fact, it is this existence of unique gene sequences that has led to one of the most unexpected results. As early as 1996, researchers were expressing surprise as analysis showed that “…approximately half of all protein coding ORFs [orphan genes] revealed by the sequence had no clearcut sequence homologs in any organism….” 2 As additional data were gathered, many of these “Open-Reading-Frames” (ORFs) or “orphan” genes still had no known function and no similar homologous sequences outside of their species or group. 2
With the advent of automated DNA sequencing in the 1990s and its growing use over the last twenty years, we now know that the genetic universe is vastly larger than previously believed. GenBank is the current database repository that stores known gene sequences drawn from hundreds of thousands of species—it is the current “dictionary” of genes that contains the specific genetic sequence and functions (if known) for hundreds of millions of different genes. These data have allowed amazing insight into the distribution of different genes across the entire spectrum of organisms. From an evolutionary perspective, these analyses are intended to compare the different genes across many species, both from very similar and different forms of life. Much like the trees of relationships inferred from different physiological characteristics, these newer data are now analyzed to discern assumed evolutionary relationships.
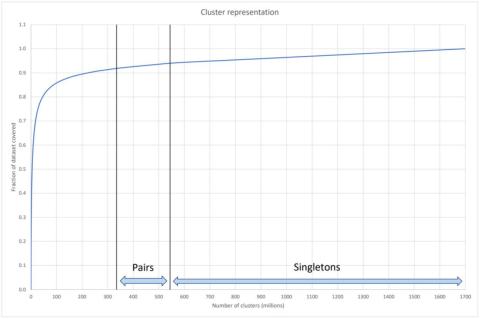
Over the last twenty years, however, it has become clear that the widespread existence of orphan genes‚—technically known as taxonomically restricted genes (TRGs)— are not a sampling anomaly but are ubiquitous across all organisms. These organism-specific genes are a fundamental aspect of the vast genomic space that defines all known protein sequences in all living organisms. Figure 2 is taken from a recent report on a new analysis capability (DeepClust) to group gene sequences in clusters to simplify analysis. This figure shows that over 1.9 billion different gene sequences were analyzed and grouped into 1.7 billion “clusters” of which over 1.1 billion sequences reside in their own clusters—they did not cluster with any other sequence. 3
Not only are these orphan genes unique for their species or group, but they are also separated in the “genetic space” by such large distances (i.e., sequence-level differences in terms of base pairs from any known other gene sequences) that it is not understood how they could have originated by any slow, incremental process from other known genes based on known genetic mechanisms.
One example of a specific study is helpful to better understand the implications of orphan genes to an evolutionary understanding of protein gene origin. Richard Buggs and co-authors published the genome of the ash tree in 2016 in Nature. They found 38,852 protein coding genes and of these 9604 (about 25%) are unique to the ash tree at either the species, family, or genus level. They report that such orphan genes are found in every new sequenced genome, and they conclude:
Orphan genes are the “hard problem” for evolutionary genomics. Because we can’t find other genes similar to them in other species, we can’t build family trees for them. We cannot hypothesise [sic] their gradual evolution; instead they seem to appear out of nowhere. Various attempts have been made at explaining their origins but—as Paul [Nelson] and I describe in our book chapter—the problem remains unsolved. 4
We see that the ubiquitous nature of these orphans or TRGs has not only been unexpected by biologists but has also created apparent conflicts that cannot be resolved within the model of UCD. If orphan genes have no close homologous genes in other organisms, where did they come from? The typical evidence for relatedness is shared genes that are assumed to indicate a flow of genetic information from an ancestor down to each modern organism. If there is no gene in any other organism that is similar enough to have evolved into the TRG, then this defies explanation under any current hypothesized evolutionary mechanism. These TRGs exist for every sequenced organism and constitute a significant percentage of the genes in each organism—they are not the exception but are the rule. Apparently, every species or group of organisms that contains TRGs is essentially “customized” to be different from all other organisms in order to carry out their functions and sustain life.
In a recent series of interviews with biologist Dr. Paul Nelson, he relates the analogy of a library full of millions of books that are to be used to create a dictionary of all the words used in the English language. As each book is examined, however, it is found that every single book contains many new words that did not occur in any other book. The dictionary would thus continue to grow, long past the point where it would be expected to converge to the set of all words that make up the English language. This unexpected result means that the books were not created using a fixed pre-existing language, but rather that each book is unique in that it contains words that do not occur in any other book in the entire library! 5
Impact on the concept of LUCA
As noted previously, LUCA is assumed by evolutionists to be the common ancestor of all living organisms—from bacteria to eucaryotes and higher animals. This is a hypothetical organism that is based on some minimal set of genes that are needed to perform the most simple and fundamental functions of life to allow assumed evolutionary processes to start the development of life on earth through naturalistic mechanisms. The necessary existence of LUCA is an assumption since it is believed that the unknown process of abiogenesis—the origination of the first living organism from nonliving material—is extremely unlikely, the common assumption is that it happened only once. Furthermore, because all organisms are descended from this LUCA without interruption, every descendent organism would necessarily need to retain those critical functions and genes that cannot be changed without compromising the ongoing process of life and evolution itself—you cannot “rebuild an airplane while it is flying.”
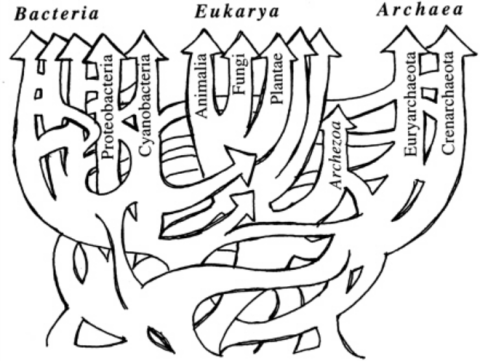
Even before the growing awareness that orphan genes are pervasive and fundamental to all organisms, there were already a growing number of biologists that rejected the idea of UCD and a single LUCA organism. Figure 3 shows a sketch of the “tree of life” (ToL) created some years ago by biologist W. Ford Doolittle that illustrates his belief that the vast array of life on earth could not be accounted for from a single common ancestor. Since that time, many biologists have expressed their skepticism that a single ToL exists and can be resolved. Multiple anomalies in the data show that the divergence of life is not consistent with UCD and therefore that there is no LUCA. 5
All such trees—even those generated by complex computer analysis—are of course not observed by data available today. Instead, they are based on mathematical extrapolation of currently observed organism genomes back in time to hypothesized “missing link” organisms with inferred genomes to somehow explain the flow,
Conclusion
With the growing understanding of the pervasiveness of TRG, it is becoming clear that gradual emergence of new genes and functions results not only in an ever-growing library of genes, but one in which the unique and divergent TRGs cannot be accounted for through evolutionary mechanisms.
Although this is still a relatively new phenomenon, awareness of the massive challenge that it poses conventional evolutionary models is growing. This and other aspects of the fundamental genetic reality are leading to more biologists questioning past assumptions and calling for a need to develop new models.
Perhaps another way to understand this new phenomenon is to instead consider that the possibility that the amazing complexity and diversity of life is the result of the purposeful activity of a Creator. A creator that apparently has made every organism with unique and distinct “fingerprints”—genes that code for species-specific proteins that cannot and did not arise through random processes of genetic drift, mutation, and recombination from previously existing genetic information and organisms, even over vast periods of time.
- 1Bergman J (2001) Does homology provide evidence of evolutionary naturalism? J Creation 15(1):26–33. https://creation.com/does-homology-provide-evidenceof-evolutionary-naturalism Accessed 2023 Sep 18
- 2 a b Dujon B (1996) The yeast genome project: what did we learn? Trends Genet. 12(7):263–270.
- 3Buchfink B, Ashkenazy H, Reuter K, Kennedy JA, Drost H-G (2023) Sensitive clustering of protein sequences at tree-of-life scale using DIAMOND DeepClust. bioRxiv (preprint: https://www.biorxiv.org/content/10.1101/2023.01.24.525373v1.full) A version posted 2023 Feb 07 was posted at the time of this writing. Accessed 2023 Sep 18
- 4Buggs R (2016 Dec 29) The evolutionary mystery of orphan genes. https://richardbuggs.com/2016/12/29/theevolutionary-mystery-of-orphan-genes/ Accessed 2023 Sep 18
- 5 a b Ahmad S (2022 Aug 17) Testing Universal Common Descent: Parts 1 to 7 - Dr Paul Nelson. https://www.youtube.com/playlist?list=PLufmopp748Z3aBGdNmRtEzQll2SFH9FRX Part 1 accessed 2023 Sep 17