Review of The Stairway to Life: An Origin of Life Reality Check
The Stairway to Life: An Origin of Life Reality Check (Fig. 1), 1 written by chemist Change Laura Tan and engineer Rob Stadler, is one of the most insightful and devastating books ever written about the problems with abiogenesis (the spontaneous formation of life from chemicals), especially the RNA world hypothesis. Written in 2020, the book is 268 pages long, is divided into three major parts and 21 chapters, has a glossary and index, is well illustrated with drawings and tables, makes well-reasoned arguments, and cites 227 articles from the scientific literature. Stairway is written clearly, concisely, and with penetrating insight into the various layers of information found in living cells, revealing the complete explanatory bankruptcy of abiogenesis. Tan (Fig. 2),
a former atheist turned Christian, 2 has training in organic chemistry, biochemistry, and genetics and is currently an associate professor of biological sciences at the University of Missouri. Stadler (Fig. 3) has training in electrical and biomedical engineering and works in the medical device industry with over 140 patents to his credit. The book discusses the various “steps” on the “stairway” from chemistry to biology (Fig. 4), citing the most recent and relevant research. After showing why chemistry and physics cannot account for a given step, the authors introduce the next step giving abiogenesis the benefit of the doubt for all previous steps. The authors bring to light many of the paradoxes and irreducible complexities in life that are inexplicable by abiogenesis. The book is a bit technical and will be most meaningful to those with backgrounds in organic chemistry, biochemistry, or molecular biology. 3
This review will discuss each chapter of the book in order.
Introduction
The authors give a brief history of the theory of spontaneous generation. People once thought that animals could spontaneously emerge from waste. Redi showed that maggots do not spontaneously emerge from rotting flesh. Later, Pasteur showed that even microscopic life does not emerge spontaneously. Life has only been observed to emerge from life. Pasteur’s work was published about the same time as Darwin’s Origin of Species. Darwin believed in abiogenesis despite Pasteur. Despite what we know from observations, abiogenesis is widely taught today.
In 2010, Craig Venter reported the first “synthetic” self-replicating cell, nicknamed Synthia. Despite all evidence to the contrary, this achievement was heralded as evidence in favor of abiogenesis. In order to build Synthia, Venter’s team borrowed 99% of Synthia’s parts from extant life, used carefully planned and executed experiments under controlled laboratory conditions, started with pure chemicals in high concentration, and employed the help of over 40 well trained scientists. 4
Tan and Stadler use the construction of Synthia throughout the book to reveal the many problems on the stairway to life for abiogenesis.
PART 1: LESSONS FROM SYNTHETIC LIFE
Chapter 1: Recipe for a Self-Replicating Cell
Tan and Stadler point out that, to date, all attempts at creating life in the lab have started with extant DNA. They discuss the Miller-Urey experiment wherein a reducing atmosphere, thought to mimic the early earth, was heated and subjected to electrical discharges to afford low yields of a few amino acids. No one has come close to demonstrating life could emerge from these conditions or even if those conditions are geochemically plausible. Venter started with existing DNA and then added a few functionless “watermarks.”
Scientists have tried to define a minimal gene set for an independent self-replicating organism. As part of this quest, over 1000 genomes of prokaryotes have been compared. Interestingly, no homologous genes were shared across all the organisms, contradicting the idea of common ancestry. Researchers use “knock-out” experiments on simple cells wherein genes are systematically removed to determine the impact on the organism. The approach has limitations. The simplest life we know of is Mycoplasma genitalium. It has 425 essential genes (as determined by knock-out experiments) which are expressed as 382 proteins and 43 RNA molecules. Science knows of no simpler viable life. Hence even the simplest cell requires hundreds of genes to survive and reproduce. Venter transplanted a slightly modified version of the genome of a related prokaryote, Mycoplasma mycoids (referred to as simply Myco from now on), into another cell to build Synthia. Hence, Venter began with an intact functional genome.
Chapter 2: Synthesis of the Synthia Genome
Venter needed to use the machinery of various organisms to build Synthia. Synthia had 1.1 million base pairs consisting of the genome of Myco (98.55%), transposons from E. coli (0.08%), a yeast vector (0.94%), and watermarks (0.43%). Synthia’s cytoplasm and membranes came from Mycoplasma capri (referred to as Capri from now on). Venter built Synthia’s genome in small pieces (with E. coli), which were subsequently joined together (in yeast). Venter started the synthesis of the genome of Synthia with pure monomers that were joined together using extant cells. Hence, he began with homochiral monomers that were polymerized using the correct functional groups. Despite these favorable conditions, there was still a 0.5% error rate, requiring periodic isolation and purification of the DNA segments before resuming the synthesis. Once the DNA segments had been joined together with E. coli into 1000 nucleotide (nt) units, Venter employed enzymes from yeast to join the 1000 nt units together. E. coli’s enzymes can replicate its genome in 40 minutes with a 10–9 error rate. If E. coli’s DNA was one meter in diameter, the enzymes that duplicate the genome would be like a FedEx truck travelling 375 mph.
E. coli (prokaryote) and yeast (eukaryote) use different promoters and different DNA replication, transcription, and translation machinery. These differences call into question if these organisms share a common ancestor.
DNA on its own can do nothing; it needs other molecules for replication, untangling, stabilization, and transcription. Hence, it could not have been the starting place for abiogenesis.
Chapter 3: Decoding the Code
The Synthia genome had to be put in the right cell with the correct machinery and code breaking capability. Hence the Synthia genome would not be functional in yeast because portions of the genetic codes employed in prokaryotes and eukaryotes are different, again calling into question common ancestry.
Messenger RNA (mRNA) needs a translation initiation site, machinery to recognize the reading frame, machinery that recognizes the start and stop codons, and a protein coding region. Every mRNA has three potential reading frames so could code for three different proteins. Starting codons vary by organism: yeast: AUG; E. coli: AUG, UUG, AUU, AUC, AUA, CUG; mycoplasmas: UUA and all pertaining to E. coli. Hence bacteria and eukaryotes differ in the way they define and recognize translation initiation sites. UGA is a stop codon for E. coli and yeast but codes for tryptophan in mycoplasmas. There are 33 different sets of codons in extant life. How could code changes take place during evolution without killing an organism? Yeast could replicate the Synthia genome but could not decode it. Only mycoplasmas have the right machinery to decode the Synthia genome. Hence the code and decoding machinery must be in place simultaneously for the system to work!
Chapter 4: So Close, Yet So Far Away
Every cell is equipped with restriction enzymes (REs) that detect and eliminate foreign DNA. The pattern of methylation of the DNA (epigenome) is what the REs recognize. Venter’s team had to transfer Synthia’s genome to a cell with the right REs. Capri was incompatible even though Synthia’s DNA is based mostly on Myco. Since most of Synthia’s DNA was made using yeast, the methylation pattern was wrong and rejected by Capri’s REs. Venter’s team solved this problem two ways. One way was to remethylate Synthia’s genome in vitro with extracted Capri enzymes so Capri’s REs would accept Synthia’s genome. The other approach was to delete the Capri RE genes.
Essential genes are genes that a cell must have to survive or reproduce; if an essential gene ceases to function, the cell dies or can’t replicate. During the synthesis of Synthia, a mistake was made in one of Synthia’s genes: one nucleotide was omitted. The omission resulted in a frame shift that lead to production of an incorrect protein. The correct protein would have bound to the origin of replication. The overall result was that this version of Synthia could not replicate. This shows how vulnerable organisms are to some mutations–yet another barrier to evolution.
Chapter 5: The Culmination of 15 Years of Work
When a properly methylated version of Synthia’s genome was placed into Capri, cell replication resulted in three cell varieties: cells with only Synthia’s genome, cells with the genomes of Capri and Synthia, and cells with only Capri’s genome. Venter’s team had added a gene for antibiotic resistance to Synthia’s genome. After treatment of the culture with the antibiotic tetracycline, only Synthia survived. The Capri cells were killed outright. The Capri/Synthia cells could not compete for resources with the Synthia cells, eventually leaving Synthia as the sole survivor. Hence Synthia emerged due to artificial selection.
For Synthia, all the functional genes came from natural sources. All novelty came from intelligent input. How then can the creation of Synthia possibly support abiogenesis?
PART 2: THE OVERLOOKED REALITY OF LIFE’S COMPLEXITY
Chapter 6: The Stairway to Life
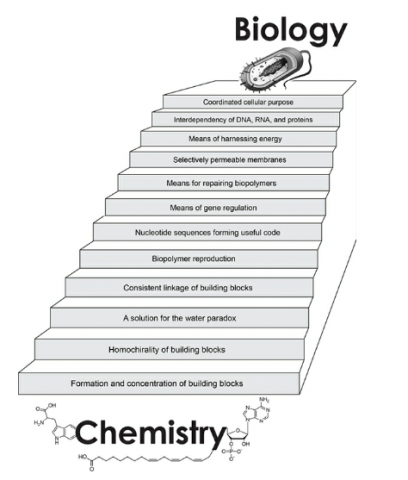
We have always underestimated the complexity of life. The stairway to life (Fig. 4) consists of 12 steps, all of which are required for viable cells. The steps are discussed individually, assuming all previous steps have been accounted for. Science can’t say with confidence how life arose. Science can, however, identify the minimum requirements for life as we know it and hence get a good idea what abiogenesis must achieve.
Chapter 7: Formation and Concentration of Building Blocks
Life’s building blocks consist of amino acids, sugars, bases, and phospholipids. Much abiogenesis research focuses on production of the building blocks using unrealistic starting materials and reaction conditions. Realistic origin of life (OOL) research into the formation of life’s building blocks requires: (1) realistic compounds are in sufficient amounts, (2) all reactions must occur in water, and (3) yields must be significant.
As mentioned earlier in the discussion of chapter 1, the famous Miller-Urey experiments used a reducing atmosphere. Miller used NH3, CH4, H2, H2O, heat, and electricity. No oxygen was permitted. The reaction products were isolated in a trap protecting them from the harmful reaction conditions. The proteinaceous compounds formed included glycine and alpha and beta alanine in low yields. Fifty years later, Bada retested some of Miller’s residues with modern equipment and repeated some of Miller’s experiments but also added H2S. Bada found 10 of the essential amino acids (racemic), but most of the reaction products were amino acids and amines not found in life.
The oxidation state of cerium found in ancient zircons strongly suggests that the atmosphere of the prebiotic earth was neutral and not reducing. OOL experiments conducted under neutral conditions afford fewer types and poorer yields of amino acids. There is little confidence that we know what the prebiotic earth was like.
Phospholipids are required to form the membranes that let molecules in and out of cells and maintain chemical and electrical gradients essential to life. Phospholipids consist of a glycerol backbone with two fatty acids and a phosphate group. Phospholipids in bacteria and Archaea are very different. The enzymes in bacteria and Archaea that form glycerol phosphate are very different. Theory says Archaea and bacteria share a common ancestor, but the data suggest otherwise. OOL experiments have made low yields of phospholipids not found in life.
The nucleotides found in RNA only have one bonding arrangement for the base, ribose, and phosphate, although hundreds are possible. There are 180 possible bonding arrangements between adenine, ribose, and phosphate, but only one is observed in life. The likelihood of polymerizing the correct version (monomer molecule with the correct bonding arrangement) from a soup containing all 180 possible versions to afford an RNA molecule with 40 nt is 180–40 or 10–90. This means it would be more likely for someone to pick a specific particle out all the particles in the universe 5 than to form an oligonucleotide made of 40 correct nucleotides by chance. And the reality is far worse because there would be other (wrong) sugars and bases in a primordial soup. Ribose is formed in low yield in OOL experiments and is not very stable. The energy sources needed to make the building blocks (heat, light, electricity, radiation, metal catalysts, etc.) can also destroy them. More time can only help abiogenesis scenarios if degradation is ignored, which is, of course, unrealistic.
Most OOL experiments produce complex mixtures with the desired products only representing a small fraction of the whole. Those who promote abiogenesis must identify mechanisms for isolating and concentrating the correct monomers. Some have suggested that the pores in the walls of hydrothermal vents could be where monomers concentrate, but it is unlikely the pores would remain stable over the millions of years needed for polymerization.
This ended the discussion of step 1 (Fig. 4).
Chapter 8: Homochirality of Building Blocks
The second step on the stairway to life is homochirality of building blocks. Chirality is a property an object possesses if the object and its mirror image are not superimposable. A good example are your left and right hands, which are mirror images but not superimposable. Molecules can also exhibit chirality. In life, all amino acids are “left-handed” while all sugars are “right-handed.” In the laboratory, the synthesis of amino acids or sugars always produces both the right- and left-handed molecules in equal amounts. No one knows how life came to have exclusively left-handed amino acids or right-handed sugars; that is, how amino acids and sugars in life are homochiral.
The drug thalidomide, a chiral molecule, was sold as a treatment for the morning sickness associated with pregnancy, and it was effective. The marketed product contained both the left- and right-handed versions of the molecule. However, it was eventually demonstrated that while the right-handed thalidomide did indeed treat morning sickness, the left-handed version caused birth defects! The point is that the three-dimensional structure of a molecule, its stereochemistry, matters.
In RNA nucleotides, there are 16 possible stereochemical forms, but only one is found in life. Those who promote abiogenesis have yet to identify a credible natural mechanism that produces only one stereochemical form or somehow filters out all the others. For proteins, even one right-handed amino acid out of hundreds of correct left-handed amino acids can destroy the protein’s function. Interestingly, the glycerol moieties of the phospholipids in bacteria and eukaryotes are exclusively left-handed, but those in Archaea are exclusively right-handed, again calling into question common ancestry. The enzymes that produce these various phospholipids all contain exclusively left-handed amino acids but are otherwise different. The amino acids in proteins can slowly become a mixture of left- and right-handed molecules (racemize), so the cell needs to keep producing new proteins. No one knows how proteins came to have only left-handed amino acids.
The best-known reaction that offers any hope for abiogenesis for the production of homochiral molecules is the Soai reaction. The Soai reaction is autocatalytic and can produce mixtures of left- and right-handed molecules with up to an 85% excess of one of the forms. However, even this excess can’t explain an exclusively left-handed amino acid world. Another problem is that the required dialkylzinc reagents are not stable in water, calling into question the relevance of the Soai reaction to understanding abiogenesis. Several other mechanisms and reactions have been suggested to explain life’s homochirality, but so far none of them are specific or efficient enough.
Chapter 9: A Solution to the Water Paradox
The authors now assumed that the basic building blocks were in hand, bringing us to the next step, which is polymerization of the building blocks in water or in the solid state to form proteins from amino acids or RNA from nucleotides. Polymerization in water is difficult because water favors the monomers over polymers. Water can also cause the monomer units in the polymers to racemize. There are some suggestions on how to overcome these problems, but they call for controlled conditions and a sequence of steps that are unrealistic for abiogenesis.
Water hydrolyzes DNA, RNA, and proteins, requiring repair mechanisms in cells. Hence repair mechanisms have to already be in place to make fresh replacement molecules. The DNA and RNA polymers can be hydrolyzed to monomers. Some of the bases can be altered or removed without breaking the polymer chain. Water is a paradox for abiogenesis: water is needed for biochemistry, but it appears to be inimical to prebiotic chemistry.
Chapter 10: Consistent Linkage of Building Blocks
The next step is to find a way to polymerize the monomers with homolinkages, that is, with the correct atoms bound together and with the correct three-dimensional structure. There are many functional groups (reactive groups of atoms) in DNA and RNA monomers which are not involved in making linkages with other monomers in the DNA and RNA polymer chains. The problem is how to get only certain functional groups to link to other monomers without participation from the other functional groups that are present. In an abiotic world, there is nothing to prevent the undesired functional groups from reacting. In life, there are enzymes that can control the polymerization so that only certain functional groups are involved in the polymerization. In an abiotic world, the only way the correct linkages can be selectively and exclusively formed is under controlled conditions designed by chemists.
In an experiment starting with pure nucleotide monomers conducted at the right temperature, pH, and concentration of monomers, only a trace of polymers with more than two monomers were formed. The largest polymer had 11 monomers (yield: 0.0003%) where one-third of the linkages were incorrect.
In another experiment conducted in the presence of montmorillonite clay, polymer chains with up to 30 monomers were formed but still with many incorrect linkages. Under these conditions, polymerization of the purine nucleotides (A, G) was favored over the pyrimidine nucleotides (C, T, U), a result that would severely limit the information capacity of DNA or RNA formed in this way. The resulting polymers were hydrolytically unstable, and the presence of clay exacerbated the problem. Some of the longer chains cyclized stopping the polymerization.
In another experiment, monomer formation and polymerization were carried out under dry conditions with proton irradiation. Polymers containing up to 20 monomers were produced. If the reaction was repeated starting with pure nucleotides, a polymer with 100 monomers with the correct linkages could be produced in low yield. Compare this result with the fact that the simplest known life has DNA with roughly 500,000 perfectly linked monomers. The experimenters started over with pure monomers because of the “asphalt paradox,” which is the observation that organic molecules left to themselves with an energy source (heat, light, etc.) form useless, complex, and intractable tars.
Proteins are formed exclusively with alpha peptide bonds although other linkages are possible. The amino acids lysine, glutamic acid, aspartic acid, and arginine can form amide linkages between monomers not found in life. An experiment wherein pure amino acids were heated together formed peptides mostly with incorrect amide linkages between the monomers.
All successful syntheses require extensive investigator intervention and are therefore irrelevant to abiogenesis. Even under these ideal controlled conditions, there are still many undesirable side reactions.
The authors summarize this section:
All empirical evidence tells us that homolinkage of DNA, RNA, and proteins can only be achieved via the highly specific and catalytic activity of enzymes and ribozymes or via the careful planning of intelligent agents. In short, without the intervention of intelligent agents, existing biopolymers are required for the production of biopolymers. 6
Chapter 11: Biopolymer Reproduction
We are now at the stage of trying to understand how the polymers of life can be faithfully replicated. In other words, we need to find a sequence of nucleotides that will result in a self-replicating RNA molecule. The RNA world hypothesis holds that the first self-replicating molecule was RNA. This idea is attractive because RNA is integral to many of the cell’s functions (e.g., mRNA, rRNA, tRNA, etc.) and the fact that some RNA molecules can promote reactions as if they were enzymes. RNA molecules that not only carry information but can also function as chemical catalysts are called ribozymes. DNA, on the other hand, can only carry information. In extant life, DNA replication in even the simplest life requires 14 complex protein enzymes and 25 polypeptides.
No self-replicating RNA molecule is known to exist in nature or to have been synthesized in a laboratory despite 50 years of effort. Nevertheless, many interesting and revealing investigations have been done to explore the possibility of an RNA world. For example, a viral RNA molecule could be replicated from pure monomers in the presence of the enzyme Qb replicase (a protein with 1200 amino acids). Allowing the system to undergo 75 replications resulted in faster replicating but smaller RNA molecules. Hence shorter RNAs with less information came to dominate, since they could replicate faster than the long, more complex, and slow-to-replicate RNA molecules. Clearly the experiment does not help abiogenesis since it required pure monomers and a complex enzyme and resulted in RNA that had less information than at the start. And, of course, this system did not involve a self-replicating RNA molecule.
There are theoretical problems for the RNA world. Let’s assume that there is a self-replicating RNA molecule that can serve as an information template and chemical catalyst. Such a system would need to start with two molecules, one to serve as a template and the other to do the chemistry. Suppose that complimentary monomers aligned with the template, and then the ribozyme stitched them together. You would now have the sense and antisense RNA strands which would strongly bind together by hydrogen bonding to form a helical dimer. The dimer would have to be unwound into its sense and antisense parts before the antisense part could serve as a template to make another sense strand and complete the replication cycle. But in the abiotic world, there would be nothing to unwind the dimer. Moreover, the longer the RNA strand, the tighter the helical dimer strands would be bound together. Experimentally, only short strands of RNA (30 nt) helical dimer, held at relatively high temperature, can be unwound and undergo more chemistry. Qb replicase worked because it is able to keep the complimentary (antisense) RNA strand separated from the sense strand.
The best attempt, to date, to demonstrate a self-replicating RNA system consisted of two RNA molecules, one with 135 ribonucleotides and the other with 153 ribonucleotides. The system required duplication of both RNA strands in order to function. So, unguided chemistry alone would have to deliver both RNA molecules at the same place and time for the system to self-replicate. The probability of doing this is slim. Even if you grant the availability of pure monomers and a way to polymerize them with the correct linkages, the probability of making even the smaller RNA molecule would be 1 out of 4135 (1.9 x 1081). This is a probability less than that for picking a specific particle out of all the particles in the universe (1080)!
Self-replicating ribozymes have other problems besides being improbable. Such a ribozyme would need to fold up into a specific three-dimensional structure in order to function as a catalyst. But if the folded structure is thermodynamically favored and the method of replication would be for complimentary nucleotides to initially align along the template before being stitched together, there would need to be some mechanism for unfolding the RNA molecule so it could serve as a template.
An interesting self-replicating RNA system that consisted of two ribozymes and four RNA ribozyme precursors has been reported, 7 but was purely man-made and hence had nothing to say about abiogenesis. 8
An RNA world faces other problems. RNA is unstable to hydrolysis. Therefore, the rate of formation of the RNA would have to be faster than its degradation or else it could never persist. The monomer ribonucleotides would need to be activated somehow (made reactive) to facilitate polymerization, but that also would mean they would be less stable. The replication process would have to operate with high fidelity or else the information required for the function of the ribozyme would be lost. Nevertheless, a small mutation rate would still be required in order for the ribozyme to evolve. After decades of research, no one has made a self-replicating RNA molecule or found one in nature.
Chapter 12: Nucleotide Sequences Forming Useful Code
Now we come to the information question. How could meaningful nucleotide sequences be formed through ordinary chemical reactions? For the RNA world to be viable, the correct sequence of ribonucleotides that would generate a self-replicating ribozyme is required; not just any old sequence of ribonucleotides will do. We know from experiment that there is no inherent tendency of ribonucleotides to polymerize in any particular order, so the only way the correct sequence could be found would be by chance.
The probability of forming a specific RNA molecule with 135 nt was discussed previously. But what if there are ten billion (1010) variants of the RNA sequence that would still work? We calculated before that the probability of forming an RNA molecule consisting of 135 ribonucleotides was 1 out of 1.9 x 1081. So, if 1010 variants were functional, the probability of forming the RNA molecule would then be one out of 1.9 x 1071 or still impossible.
Now, for the sake of argument, assume that a self-replicating RNA molecule can be formed naturally and is in hand. This self-replicating RNA molecule must now evolve into extant biochemistry through random mutations and natural selection. Theoretically, this could proceed through multiple RNA molecules each having a different function or through a single RNA molecule having multiple functions. As we have already discussed, experiments have shown that replicating RNA with enzymes results in faster replicating and smaller RNA molecules. This disfavors the scenario with a single RNA evolving into a more and more complex molecule with more and more functions. In a scenario with multiple competing RNA molecules, it is likely one would prevail rather than having several “share” resources.
What is the simplest possible life form? The simplest version of Synthia had 513,000 DNA nucleotides and 473 genes. However, this version required a very favorable environment and could not survive in the wild. The fewer the genes, the more vulnerable the organism.
Chapter 13: Means of Gene Regulation
Now, assuming a set of self-replicating RNA molecules is in hand, a way to regulate the RNA “genes” must be found. Which RNA variants survive will depend on the relative rates of replication, the tolerance to changing conditions, overall stability, etc. A way must be found for the RNA replicators to function cooperatively instead of competitively. Feedback mechanisms that control when a given RNA gene replicates and when it stops must be put into place. These feedback mechanisms would need to have a sensor that could detect gene products, a mechanism that “decided” when a critical amount of gene products had been reached, and a means for starting and stopping gene expression. These three capabilities must be integrated and coordinated.
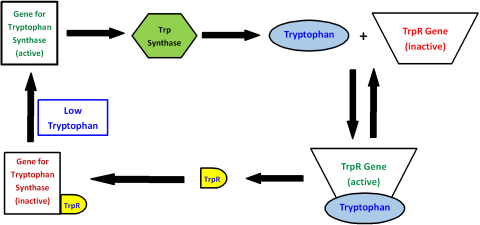
The authors use the regulation of the synthesis of the amino acid tryptophan to illustrate (Fig. 5).
Tryptophan is produced by the enzyme tryptophan synthase. As long as the gene for tryptophan synthase is active, tryptophan is produced. When the concentration of tryptophan becomes high enough, tryptophan binds to the tryptophan repressor gene (trpR). This binding activates the trpR gene, which then produces the tryptophan repressor protein (trpR). Then trpR binds to the gene for tryptophan synthase, thereby deactivating it and stopping the production of tryptophan synthase and hence tryptophan. When the level of tryptophan drops low enough, the trpR gene becomes inactive, less trpR is made, and eventually the gene of tryptophan synthase becomes active again. All of this control is in place for just one molecule. In a living cell, there are thousands of molecules which must be regulated simultaneously through similar feedback interactions.
Many complex and cooperative RNAs would be needed to establish a regulatory network.
Another real-life example is the gene glmS found in a species of bacteria. The mRNA produced by glmS is joined with an additional short segment of RNA (75 nt) that has a regulatory function. glmS codes for an enzyme. When the metabolic products of the enzyme build up, the short regulatory RNA segment detects it, changes shape, and then destroys the coding RNA, preventing further production of the enzyme and its metabolic product.
Then there is the methyltransferase (MT)/endonuclease (EN) protection system that keeps foreign DNA out of a cell. MT is an enzyme that methylates native DNA at very specific locations. EN is an enzyme that cleaves DNA at the same locations except when methylated. EN will cleave any unmarked DNA. Both MT and EN must be in place simultaneously to add any value to the cell. What could have been the step by step evolutionary pathway to this system? How could cells have survived without the system?
In an RNA world, smaller and faster replicating RNA strands would outcompete the larger, more slowly replicating RNA strands for limited resources making it difficult to produce molecules with large information carrying capacity. Only with a regulatory system where smaller RNA molecules were labeled and then destroyed could the larger RNA molecules thrive. The complete regulatory system would need to be in place to provide any benefit.
Chapter 14: Means of Repairing Biopolymers
DNA in extant cells is constantly degrading via a myriad of mechanisms: hydrolysis, oxidation, radiation damage, alkylation, single strand cleavage, double strand cleavage, free radicals, copying errors, and more. However, extant cells are equipped with several repair mechanisms. There is a repair system in place in both prokaryotes and eukaryotes that can replace an excised base along DNA. The system consists of five different enzymes.
If DNA repair mechanisms fail, embryonic death may occur. If the embryo survives, the child may suffer from various maladies such as premature aging, growth retardation, and an increased risk of cancer.
There are many errors that occur during DNA replication. There are three types of DNA polymerases. Each can detect and correct errors (perform proofreading). In bacteria, proofreading decreases the DNA replication error rate by a factor of 100.
The protein molecules that carry out proofreading, and hence perform DNA repair, are coded for in DNA. How could DNA have evolved without the repair mechanism? How could the DNA repair mechanism have evolved without the DNA repair mechanism? This is a serious problem for the RNA world because RNA is even less stable than DNA. How could an RNA world have evolved without a repair mechanism?
This leads us to Eigen’s Paradox. The longer an RNA molecule becomes, the more errors it will accumulate. After several generations without repair, the accumulated error load would lead to error catastrophe and the system would cease to function. This reality limits the sustainable length a replicating RNA molecule could have. Hence replicating RNA requires repair mechanisms which need more RNA (to code for the repair mechanisms) than the error rate will allow. As the authors observe:
What we need for abiogenesis is a self-replicating RNA that has just the right error rate during reproduction, resulting in information gain over millions of years with a replication rate that somehow improves over time and the preservation of information via repair mechanisms that spontaneously appear. What we have is the absence of a self-replicating molecule, evidence that replication processes prefer shortening and simplification of RNA over time, known instability of RNA over time, and no hope of arriving at the required repair mechanisms. 9
Chapter 15: Selectively Permeable Membranes
All cells have an outer bilayer membrane consisting of phospholipids. Phospholipids spontaneously form bilayer spheres (vesicles) or sheets in water. Extant cell membranes selectively allow in molecules that provide food, building materials, and energy, and keep out waste and harmful molecules. The control is mediated by pores, consisting of special proteins embedded in the membrane, that selectively open and close in response to the detection of various types of molecules. Cell membranes also facilitate intercellular communication. Cell membranes must be able to sustain a proton gradient in order to power various molecular machines embedded in the membrane (electric motors!). Even Mycoplasma genitalium has 140 different types of proteins embedded in its membrane. One third of all known proteins function in cell membranes. Some pores are able to let water in or out of the cell without transporting protons.
One of the pores in the E. coli cell membrane is called the ABC transporter. The ABC transporter allows a molecule designated as MccJ25 to exit the cell. Membrane pores can have two gates, one on the inside of the membrane and the other on the outside. The MccJ25 enters the open pore from the inside of the cell. The closing of the inside pore and opening of the outside pore is then powered by two ATP molecules resulting in the release of MccJ25 to the outside. Another ATP molecule then powers the closure of the outer pore and opening of the inner pore to set the stage for another cycle. MccJ25 blocks RNA polymerase (needed for transcription) from functioning. When E. coli is starving, it releases MccJ25 to stop the use of precious resources by other cells.
The outer cell membrane requires embedded proteins (pores) to have the semipermeable functionality. The semipermeable function is also needed in the membrane of mitochondria in order to produce the ATP which powers the opening and closing of pores (more on this below). In other words, you need ATP to power a semipermeable membrane but you need the semipermeable membrane to produce ATP. Both must be in place simultaneously or the semipermeable membrane will not function.
Several proteins are required to facilitate the insertion of proteins into the cell membrane through complex step by step processes. The locations of insertion are marked by proteins in the membrane. These locations are in inherited from the mother cell and have nothing to do with DNA.
Chapter 16: Means of Harnessing Energy
Energy must be constantly applied to maintain order in the cell. The cell’s energy source is a proton gradient held across cell membranes. The proton gradient is used to produce ATP, the energy currency of the cell. ATP can be thought of as “life’s battery.” ATP is used to power numerous activities within the cell: activation of amino acids for protein synthesis, DNA replication, untangling DNA, breaking chemical bonds, transporting molecules, etc. Humans regenerate their weight in ATP daily. The harnessing and storage of energy in the cell is called chemiosmotic coupling (CC). CC requires a semipermeable membrane, a means of pumping protons across the membrane, and a mechanism to produce ATP. These three requirements involve complex interdependent molecular machines. Similar mechanisms for harnessing and storing energy are found in all life.
Energy from food is used to generate the proton gradient. The mitochondria generate ATP. Mitochondria are organelles that have a semipermeable membrane. An electric current is generated from the oxidation of food molecules. The current passes through the electron transport chain (embedded in the mitochondrial membrane), ultimately reducing oxygen to water and producing carbon dioxide. The current powers proton pumps in the membrane of mitochondria that move protons from outside to inside the mitochondrial membrane. Hence more protons end up inside the mitochondria than outside, establishing the proton gradient. The excess protons in the mitochondria then power a molecular machine embedded in the mitochondrial membrane called ATP synthase, which generates ATP that is released into the cytoplasm where it is available to the rest of the cell to meet its energy needs. Protons move from inside the mitochondria, through ATP synthase (giving it power), back to the outside.
The electron transport mechanism consists of four protein complexes embedded in the mitochondrial membrane, three of which will be discussed: complexes I, III, and IV. The complexes pump protons into the mitochondria. Complex I consists of between 16 and 44 proteins, depending upon species. These proteins all have hundreds of amino acids. Complex III consists of 3 to 11 proteins while complex IV has 6 to 14 proteins. In mitochondria, a potential difference of 0.2 V (volts) is maintained across the membrane. This is equivalent to an electric field of 3 x 107 V/m2, similar in strength to a bolt of lightning!
The authors refer to ATP synthase as an engineering “masterpiece.” 10
It consists of at least 20 interconnected proteins assembled in eight unique subunits. The ATP synthase operates at 7800 rpm. Each rotation, powered by 10 protons moving through the ATP synthase from inside to outside the mitochondrial membrane, generates three ATP molecules.
Chemiosmotic coupling requires at least 45 complex proteins. So, what is the evolutionary explanation for these wonders? From the Stock Lab video illustrating the ATP synthase machine, Nick Lane comments:
The standard mechanisms of evolution eventually produced the sophisticated proteins in early cells, including ribosomes and the ATP synthase, proteins conserved universally across life today. 11
In other words, we have no idea! Nobody has an explanation for how chemiosmotic coupling came to be!
Chapter 17: Interdependency of DNA, RNA, and Proteins
We will now assume that self-replicating RNA genes that are controlled by and code for well-regulated metabolic systems, powered by chemiosmotic coupling and an ATP energy economy, all housed in semipermeable membranes, are in hand. RNA repair mechanisms are in place. This is the zenith of the RNA world where ribozymes dominate. Of course, no such life form has ever been created or observed.
The minimal number of genes required for life has been estimated to be 493. An RNA world can’t provide this because ribozymes don’t have the needed versatility that protein enzymes have. The problem now before us is how to move from an RNA world to a world where DNA is used for information storage and proteins are used as enzymes.
The sequence of ribonucleotides in a self-replicating RNA molecule provides the information required for the molecule to assume the correct three-dimensional structure that enables its catalytic capabilities. However, there is no reason to believe that this sequence of ribonucleotides would have anything to do with the required sequence of ribonucleotides in an mRNA molecule that coded for a protein enzyme with the same function. Hence the RNA world, if it ever existed, would be a dead end insofar as a clear path to extant life. Hence in order to have the needed sequences of nucleotides for extant life, we would have to start over! And even if by some miracle the information in RNA-world genes did correspond to what is needed to build proteins with similar functions, you would still need to account for ribosomes and DNA transcription and replication. DNA can’t be replicated without enzymes, and enzymes can’t be built without the correct information in DNA and the genetic code.
For the transcription of DNA into mRNA, you need RNA polymerase (which has four protein subunits in prokaryotes) and promoter and stop codons on the DNA. There is no transcription without RNA polymerase and no RNA polymerase without DNA that codes for its amino acid sequence.
The ribosome is the molecular machine where the ribonucleotide sequence in mRNA is translated into an amino acid sequence for a protein. In prokaryotes, the ribosome consists of 48 proteins and three RNA molecules, all folded and assembled correctly to facilitate function. The proteins of the ribosome are coded for in DNA but could have only been manufactured by a ribosome.
Where did the genetic code come from? Some have referred to it as a “frozen accident” because any changes to it would be lethal. Interestingly, there are 33 known distinct codes. In an RNA world, what would have been the selective pressure to produce a complex translation system before there were proteins?
There are many proteins required to manage and utilize the information in DNA. They are needed for transcription, replication, unwinding, winding, compacting, repairing, etc. The molecules of the DNA/RNA/protein world are all interdependent. None can function without the others.
Chapter 18: Coordinated Cellular Purpose
It is not enough to have all the correct parts of a cell. All the functional parts must be assembled correctly to function properly. Another message of this chapter is that the central dogma (DNA makes RNA makes proteins makes us) is incomplete because it ignores epigenetic information.
Chapter 19: The View from the Top of the Stairway
The path to life from the top of the stairway looks more like a cliff. This is because most of the steps must be crossed at once for life as we know it to function. According to abiogenesis, as soon as molecules are needed, they appear. The mechanisms to repair and replace molecules are needed as soon as a self-replicating system appears, otherwise fragile information carrying molecules and catalysts will degrade and cease to function. Information in DNA is useless unless there also exists a genetic code, decoding mechanism, and the machinery to replicate DNA. A semipermeable membrane is also needed immediately, or the molecular system will diffuse apart. Those who believe in abiogenesis look to an old universe with many earth-like planets to improve the odds, but the stairway to life is really a cliff that must be scaled rapidly, so more time does not help. The next step to improve the odds is to invoke the multiverse, something there is no evidence for and can’t be falsified.
Synthia was built by trained chemist using pure starting materials and parts borrowed from extant life–all irrelevant to abiogenesis.
PART III: CONFLICT RESOLUTION
Chapter 20: Why Do People Believe in Abiogenesis
In Part III, the discussion turns more to philosophical issues. The authors discuss methodological naturalism which says experimental science must assume natural causes for all observable phenomena. Methodological naturalism has nothing to say about the existence of God because He is out of scope. Methodological naturalism has its limits and these limits need to be recognized. Then there is philosophical or metaphysical naturalism which holds that natural causes are all that exist (scientism). Methodological naturalism is a philosophy for conducting science while metaphysical naturalism is an all-inclusive worldview. If one adopts the philosophical naturalistic (atheistic) view, abiogenesis is the only possible explanation for life since God is excluded from consideration by definition. In this view, methodological naturalism must be able to explain everything since there are only natural causes. Public schools are not allowed to present the problems with evolution, resulting in the promotion of philosophical naturalism and the alleged conflict between science and religion. Those that hold to philosophical naturalism have faith that all phenomena are reducible to chemistry and physics.
Chapter 21: Supplanting Abiogenesis
Evolutionists have painted themselves into a corner: they insist methodological naturalism is the only correct epistemology for understanding origins despite the impossibility of abiogenesis. Those that believe in abiogenesis think there is an as yet undiscovered physical law that can explain the emergence of life. They have faith in naturalism.
In my opinion (Dan Reynolds), science has disproved abiogenesis. Science has explored the possible material explanations for life and has found none. The only known source of the type of information found in life is intelligence. Instead of assuming a rigid methodological naturalism insofar as origins is concerned, science needs to be able to make inferences to the best explanation without the a priori exclusion of competing possible explanations. To do otherwise will commit science to futile investigations that keep giving the same answer: chemistry and physics did not invent biochemistry.
The authors also included an epilog and a brief section called “anticipated objections” which I will not discuss here.
Summary
In summary, the authors did a masterful job of demonstrating many of the problems for abiogenesis. These problems include the complex and intractable mixtures obtained from prebiotic chemistry experiments, the problem of homochirality, the need for water yet the hydrolytic instability of life’s monomers and polymers, the problem of homolinkages, the need for a sequence of nucleotides that would create a self-replicating molecule, the need for the coexistence of various self-replicating molecules that worked together cooperatively instead of competitively through regulatory mechanisms, the need for repair mechanisms that operate faster than degradation processes, the need for semipermeable membranes for controlling molecular traffic into and out of the cell and to maintain proton gradients for powering molecular machines embedded in the membrane, the need for a means to harness and store energy, a means for moving from the RNA world to the DNA/RNA/protein world with complex enzymes and molecular machines made out of proteins, and the correct arrangement of cellular parts for the a cell to be viable.
One of the deeper insights concerning the RNA world is how it would lead to a dead end insofar as explaining the extant DNA/RNA/protein world. There would likely be no correspondence between the sequence of ribonucleotides required for a self-replicating ribozyme and the required nucleotide sequence of an mRNA molecule that coded for a protein with a similar function. Also insightful was the identification of the many molecules and functions that would need to be present at the same time. For example, as soon as a potentially self-replicating RNA molecule emerged, there would have to be already in place:
- A supply of pure monomers
- Repair mechanisms to prevent the system from ceasing to function due to error catastrophe and degradation
- Molecules that could keep the sense and antisense RNA strands separated to facilitate the replication cycle
- Molecules that could identify and destroy smaller and faster replicating RNA molecules so that larger RNA molecules with greater information capacities could survive and dominate
- Some type of containment system such as a cell membrane to prevent diffusion and dilution of monomers, etc.
Only the foresight of an intelligence could anticipate these needs; blind chance, chemistry, and physics have no foresight. Understanding the many molecules and functions required for life reveals life’s irreducible complexity and staggering improbability. Since a self-replicating RNA molecule has never been observed in nature or produced by well-trained chemists in over 50 years of trying, why should anyone suppose the RNA world is a realistic explanation for the origin of life? Clearly, nature is not up to the task of creating life, only maintaining it. Only a Brilliant Intelligence could have made us. The intricacy of biochemistry unmistakably points to a Creator (Rom. 1:19–20).
- 1Tan CL, Stadler R (2020) The Stairway to Life: An Origin-of-Life Reality Check, Evorevo Books.
- 2Tan gives some of her testimony and an overview of Stairway in a YouTube video: https://www.youtube.com/watch?time_continue=78&v=8of-OgJ3qh8&feature=emb_logo
- 3Nevertheless, the book is accessible to anyone willing to do their homework. There is a free introductory biochemistry text on the web for those who want a reference: https://biochem.oregonstate.edu/sites/biochem.oregonstate.edu/files/ahern/Biochemistry%20Free%20For%20All%201.3_compressed.pdf
- 4James Tour, one of the world’s leading organic chemists, comments on Venter’s work. See James Tour on the Riddle of Life's Beginnings (Science Uprising EP5) https://www.youtube.com/watch?time_continue=1&v=Ymjlrw6GmKU&feature=emb_logo Accessed 2020 Jun 21
- 5It is estimated that there 10 80 particles in the universe.
- 6Tan CL, Stadler R (2020) 105
- 7Lincoln TA, Joyce GF (2009) Self-sustained replication of an RNA enzyme, Science 323: 1229–1232 Reynolds D (2009 Jun) Has science found how life began
- 8Reynolds D (2009 Jun) Has science found how life began and species evolved? An examination of the “RNA World” hypothesis and rapidly changing lizards. https://tasc-creationscience.org/newsletter/june-2009-tasc-newsletter Accessed 2020 Jun 21
- 9Tan CL, Stadler R (2020) 139–140
- 10Stock Lab, The Victor Chang Cardiac Research Institute (2016 Apr 14) “ATP Synthase,” https://www.youtube.com/watch?v=b_cp8MsnZFA Accessed 2020 Jun 15
- 11Tan CL, Stadler R (2020) 159